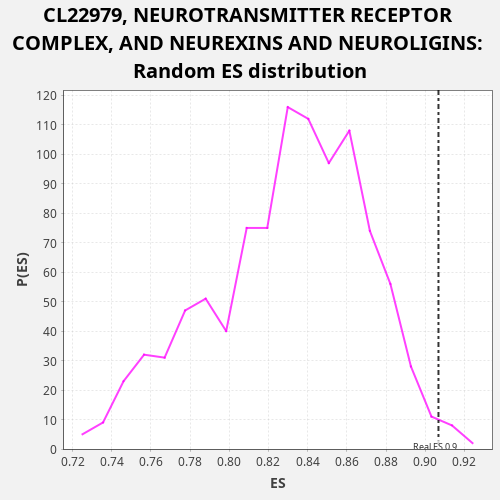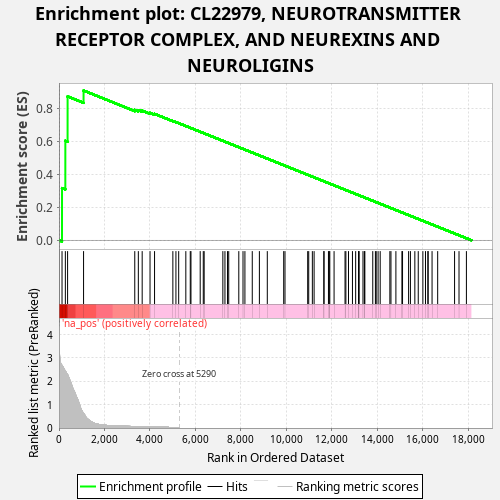
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Alopecia custom__Alopecia_custom-both_sexes- |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_pos |
| GeneSet | CL22979, NEUROTRANSMITTER RECEPTOR COMPLEX, AND NEUREXINS AND NEUROLIGINS |
| Enrichment Score (ES) | 0.90678626 |
| Normalized Enrichment Score (NES) | 1.0924326 |
| Nominal p-value | 0.014 |
| FDR q-value | 0.4042072 |
| FWER p-Value | 0.964 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | CLSTN3 | 133 | 2.670 | 0.3156 | Yes |
| 2 | GRIN2C | 279 | 2.434 | 0.6020 | Yes |
| 3 | DLG2 | 378 | 2.271 | 0.8712 | Yes |
| 4 | GRIN3B | 1080 | 0.615 | 0.9068 | Yes |
| 5 | HTR3C | 3331 | 0.060 | 0.7895 | No |
| 6 | GRIK1 | 3488 | 0.057 | 0.7878 | No |
| 7 | FBXO40 | 3657 | 0.053 | 0.7849 | No |
| 8 | HTR3D | 4002 | 0.046 | 0.7714 | No |
| 9 | TJAP1 | 4201 | 0.043 | 0.7656 | No |
| 10 | HOMER1 | 5006 | 0.009 | 0.7222 | No |
| 11 | NLGN4X | 5141 | 0.006 | 0.7155 | No |
| 12 | LHFPL4 | 5262 | 0.003 | 0.7092 | No |
| 13 | NRXN3 | 5576 | -0.000 | 0.6919 | No |
| 14 | HOMER3 | 5771 | -0.000 | 0.6811 | No |
| 15 | CBLN4 | 5802 | -0.000 | 0.6795 | No |
| 16 | CACNG5 | 6208 | -0.000 | 0.6570 | No |
| 17 | DLGAP4 | 6343 | -0.000 | 0.6496 | No |
| 18 | DLG3 | 6382 | -0.000 | 0.6475 | No |
| 19 | CBLN1 | 7204 | -0.000 | 0.6021 | No |
| 20 | HOMER2 | 7296 | -0.000 | 0.5970 | No |
| 21 | OLFM2 | 7408 | -0.000 | 0.5909 | No |
| 22 | GRIN2D | 7462 | -0.000 | 0.5879 | No |
| 23 | NRXN2 | 7905 | -0.000 | 0.5635 | No |
| 24 | MDGA1 | 8090 | -0.000 | 0.5533 | No |
| 25 | CDH9 | 8174 | -0.000 | 0.5487 | No |
| 26 | DLGAP3 | 8497 | -0.000 | 0.5308 | No |
| 27 | GRIA2 | 8810 | -0.000 | 0.5136 | No |
| 28 | GRM4 | 9158 | -0.000 | 0.4944 | No |
| 29 | DLG4 | 9879 | -0.000 | 0.4545 | No |
| 30 | CHRM3 | 9936 | -0.000 | 0.4514 | No |
| 31 | CBLN2 | 10934 | -0.000 | 0.3962 | No |
| 32 | CACNG8 | 10979 | -0.000 | 0.3937 | No |
| 33 | CNIH3 | 11139 | -0.000 | 0.3849 | No |
| 34 | CNTN4 | 11216 | -0.000 | 0.3807 | No |
| 35 | HTR3B | 11645 | -0.000 | 0.3570 | No |
| 36 | GRIK4 | 11651 | -0.000 | 0.3567 | No |
| 37 | GRID2 | 11847 | -0.000 | 0.3459 | No |
| 38 | GRIA4 | 11880 | -0.000 | 0.3442 | No |
| 39 | GRM1 | 11906 | -0.000 | 0.3428 | No |
| 40 | GRIA1 | 12094 | -0.000 | 0.3324 | No |
| 41 | SHANK1 | 12576 | -0.000 | 0.3058 | No |
| 42 | SHANK2 | 12613 | -0.000 | 0.3038 | No |
| 43 | LRRTM1 | 12728 | -0.000 | 0.2975 | No |
| 44 | GRIK3 | 12902 | -0.000 | 0.2879 | No |
| 45 | GRIK2 | 13043 | -0.000 | 0.2802 | No |
| 46 | PTCHD1 | 13172 | -0.000 | 0.2731 | No |
| 47 | NLGN4Y | 13182 | -0.000 | 0.2726 | No |
| 48 | SLITRK5 | 13189 | -0.000 | 0.2722 | No |
| 49 | NETO1 | 13365 | -0.000 | 0.2625 | No |
| 50 | HTR3A | 13428 | -0.000 | 0.2591 | No |
| 51 | CACNG2 | 13447 | -0.000 | 0.2581 | No |
| 52 | GRM5 | 13795 | -0.000 | 0.2389 | No |
| 53 | NLGN1 | 13911 | -0.000 | 0.2325 | No |
| 54 | NLGN2 | 13946 | -0.000 | 0.2306 | No |
| 55 | DLGAP1 | 14029 | -0.000 | 0.2261 | No |
| 56 | NETO2 | 14122 | -0.000 | 0.2210 | No |
| 57 | VWC2L | 14547 | -0.000 | 0.1975 | No |
| 58 | CNIH2 | 14595 | -0.000 | 0.1949 | No |
| 59 | GRIN1 | 14812 | -0.000 | 0.1830 | No |
| 60 | GRM8 | 15077 | -0.000 | 0.1683 | No |
| 61 | NRXN1 | 15104 | -0.000 | 0.1669 | No |
| 62 | GRID1 | 15371 | -0.000 | 0.1522 | No |
| 63 | GRIN2A | 15451 | -0.000 | 0.1478 | No |
| 64 | CHRM5 | 15645 | -0.000 | 0.1371 | No |
| 65 | HTR3E | 15795 | -0.000 | 0.1289 | No |
| 66 | SHISA7 | 15998 | -0.000 | 0.1177 | No |
| 67 | SHISA6 | 16113 | -0.000 | 0.1114 | No |
| 68 | GRM7 | 16222 | -0.000 | 0.1054 | No |
| 69 | NLGN3 | 16226 | -0.000 | 0.1052 | No |
| 70 | SYNGAP1 | 16402 | -0.000 | 0.0955 | No |
| 71 | GRIN3A | 16649 | -0.000 | 0.0819 | No |
| 72 | SHISA9 | 17392 | -0.000 | 0.0408 | No |
| 73 | SHANK3 | 17587 | -0.000 | 0.0301 | No |
| 74 | GRIN2B | 17912 | -0.000 | 0.0121 | No |